Mutation structure preparation
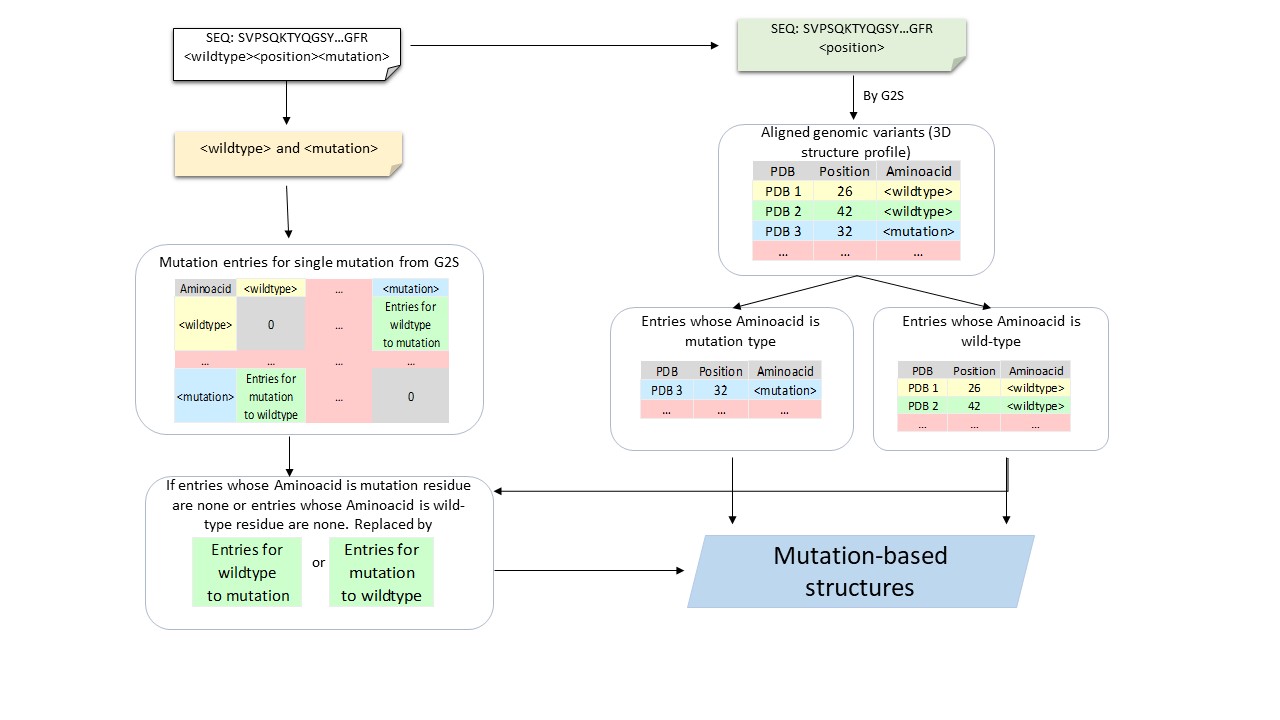
Our in-house [G2S](https://g2s.genomenexus.org) provides a real-time web Application Programming Interface (API) that automatically maps genomic variants on 3D protein structures. Giving a protein sequence and the position of a variant as the query, G2S searches similar sequence fragments (covering the surrounding regions of the mutation) in PDB to get a 3D structure profile from a list of protein structures with similar local sequences. G2S then chooses protein structures containing either wild-type amino acid or mutant amino acid at the aligned position of the mutant (queried residue; Supplementary Figure S2). According to the availability of PDB structures, four different strategies (Q1-Q4) may be adopted (Figure 2A). Q1: Tertiary structures of the wild-type residue and mutant residue are available. Q2: Tertiary structures of the wild-type residue are available, and tertiary structures of the mutant residue are unavailable. Q3: Tertiary structures of the wild-type residue are unavailable, and tertiary structures of the mutant residue are available. Q4: Neither tertiary structures of the wild-type residue nor tertiary structures of the mutant residue are available.
The samples count for 4 situations when mutation-based structures are available or not on datasets S1676, S543 and S236.
Dataset |
Q1 |
Q2 |
Q3 |
Q4 |
|---|---|---|---|---|
S543 |
172 |
366 |
1 |
4 |
S236 |
46 |
159 |
0 |
31 |
S1676 |
706 |
950 |
0 |
20 |