Casestudy
Predicting the impact of single-residue mutations on p53 thermodynamic stability
To further demonstrate the applicative power of MU3DSP, we applied it to a disease-related protein p53 containing 42 mutations. The mutations include 31 stabilizing mutations and 11 destabilizing mutations. p53 case datasets can be downloaded from p53.
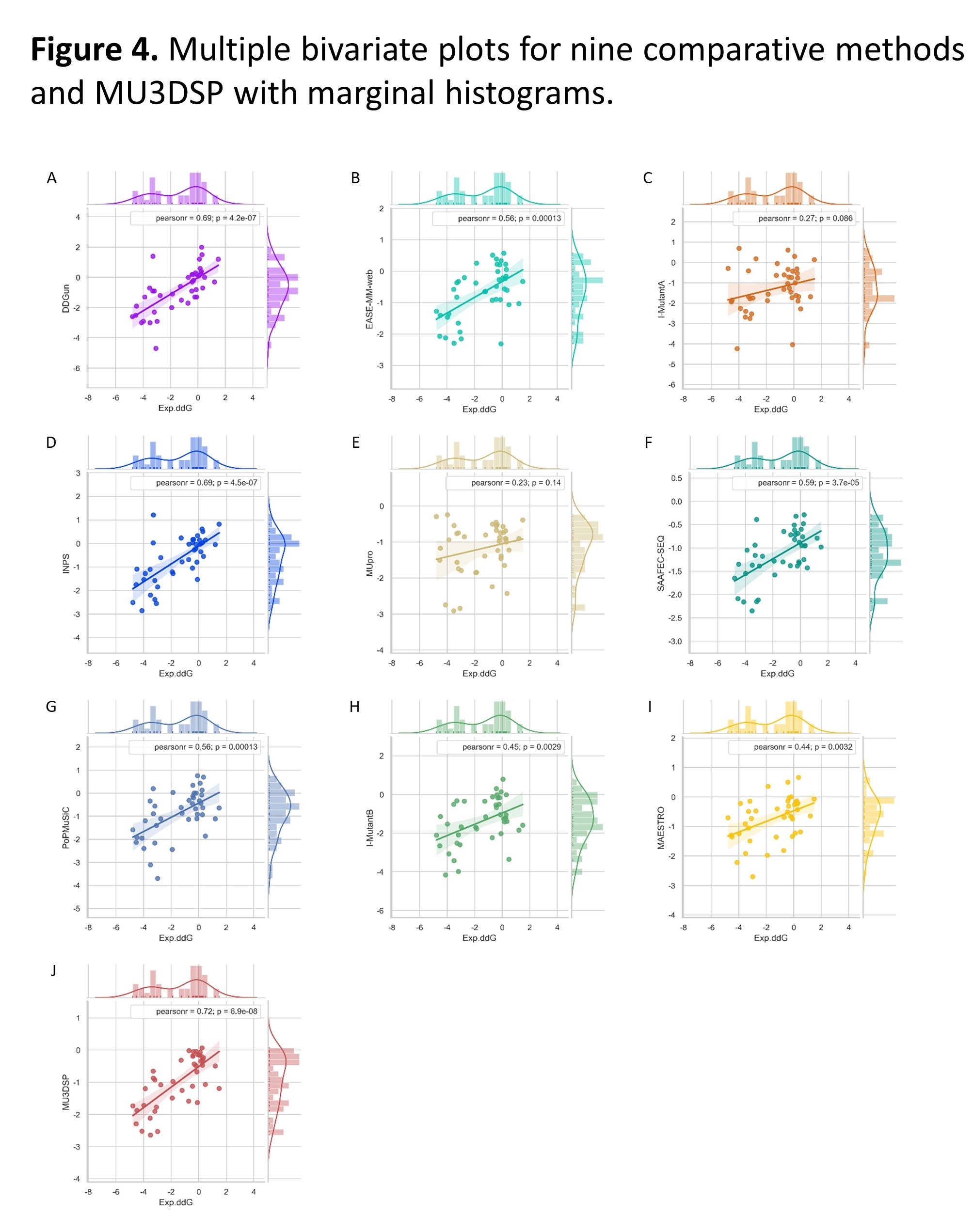
Multiple bivariate plots for nine comparative methods and MU3DSP with marginal histograms. DDG predicted with the three structure-based methods (G, H, I) and six sequence-based methods (A, B, C, D, E, F) including MU3DSP as function of experimentally measured stability changes (Exp.ddG) from the p53 dataset. The lines are the linear regression fits. Pearsonr represents Pearson correlation coefficient.
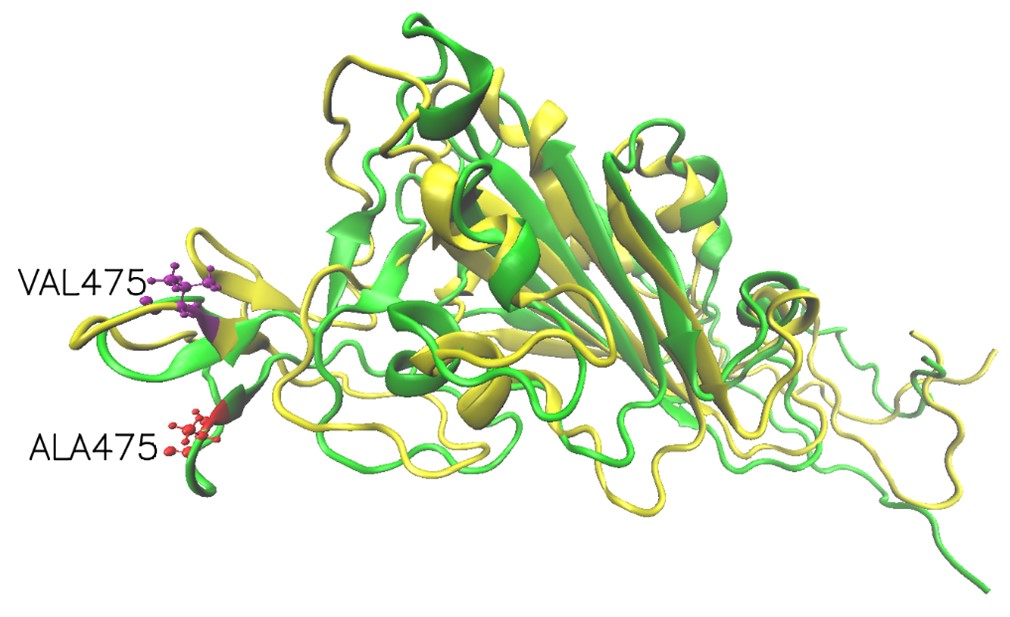
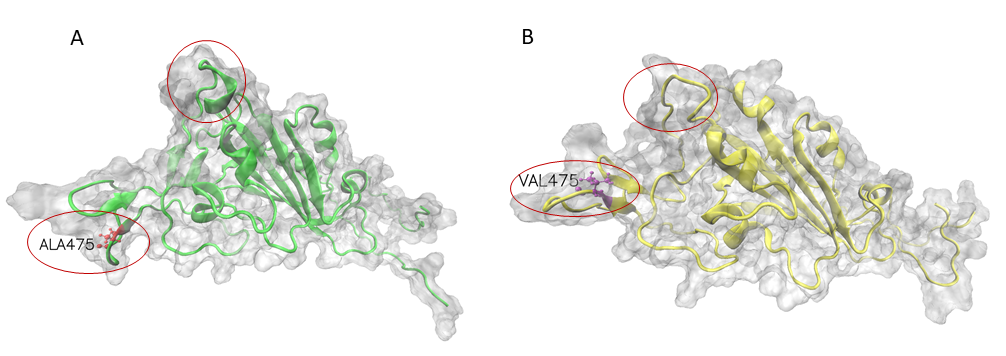
Mutation effects on for SARS-CoV-2 variants by stability changes perspective
The S proteins form homo-trimers on the virus surface and are necessary for the entrance of the virus into the host. Here, we only explored the effects of mutation on monomeric spike protein. The receptor-binding domain (RBD) is from 319 to 541 (UniProt ID: P0DTC2). Due to three PDB structures i.e. 6VXX (Closed state), 6VYB (Open state) and 6VSB (Prefusion) being discontinuous, we try to demonstrate wild-type and mutation RBD of spike protein 3D structures by AlphaFold2. The structures had visible changes after the A475V mutation and their structures align shown as follows.